Publications
This list includes only the lab’s main publications. For a more complete overview with co-authored
publications and citation data, please see Christoph Bock’s Google Scholar profile.
* corresponding author; # shared first / shared last author

Multi-omics analysis of innate and adaptive responses to BCG vaccination reveals epigenetic cell states that predict trained immunity
Moorlag SJCFM#, Folkman L#, ter Horst R#, Krausgruber T#, Barreca D, Schuster LC, Fife V, Matzaraki V, Li W, Reichl S, Mourits VP, Koeken VACM, de Bree CJ, Dijkstra H, Lemmers H, van Cranenbroek B, van Rijssen E, Koenen HJP, Jossten I, Xu CJ, Li Y, Joosten LAB, van Crevel R, Netea M#*, Bock C#*
Immunity 57/1, 171-187 (2024). DOI: 10.1016/j.immuni.2023.12.005
Donwload PDF
Read online (without subscription)
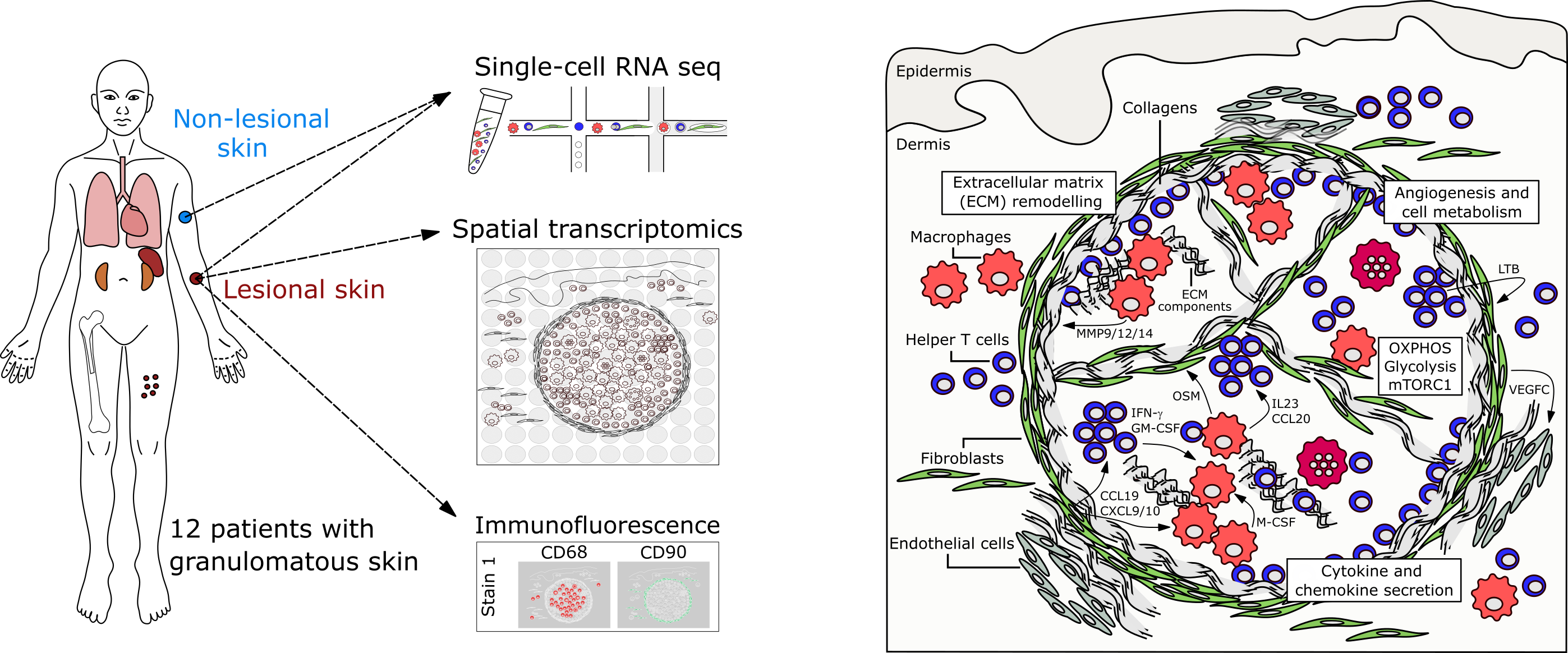
Single-cell and spatial transcriptomics reveal aberrant lymphoid developmental programs driving granuloma formation
Krausgruber T, Redl A#, Barreca D#, Doberer K, Romanovskaia D, Dobnikar L, Guarini M, Unterluggauer L, Kleissl L, Atzmüller D, Mayerhofer C, Kopf A, Saluzzo S, Lim CX, Rexie P, Weichhart T, Bock C*, Stary G*
Immunity 56, 289-306 (2023). DOI: 10.1016/j.immuni.2023.01.014
Donwload PDF
Read online (without subscription)
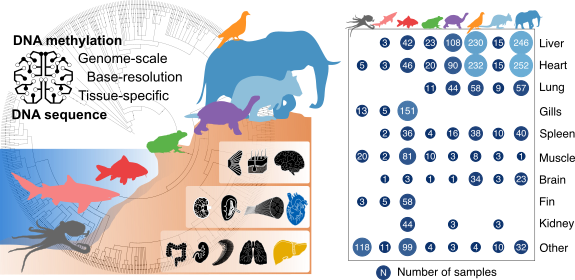
Comparative analysis of genome-scale, base-resolution DNA methylation profiles across 580 animal species
Klughammer J, Romanovskaia D#, Nemc A, Posautz A, Seid C, Linda, Schuster, Keinath M, Ramos JSL, Kosak L, Evankow A, Prinz D, Kirchberger S, Datlinger P, Fortelny N, Schmidl C, Farlik M, Kaja, Skjærven, Bergthaler A, Liedvogel M, Thaller D, Burger PA, Hermann0 M, Distel M, Distel DL, Kübber-Heiss A, Bock C
Nature Communications 14, 232 (2023). DOI: 10.1038/s41467-022-34828-y
Donwload PDF
Read online (without subscription)

LIQUORICE: detection of epigenetic signatures in liquid biopsies based on whole-genome sequencing data
Peneder P, Bock C, Tomazou EM*
Bioinformatics Advances 2 (2022). DOI: 10.1093/bioadv/vbac017
Donwload PDF
Read online (without subscription)
Press release
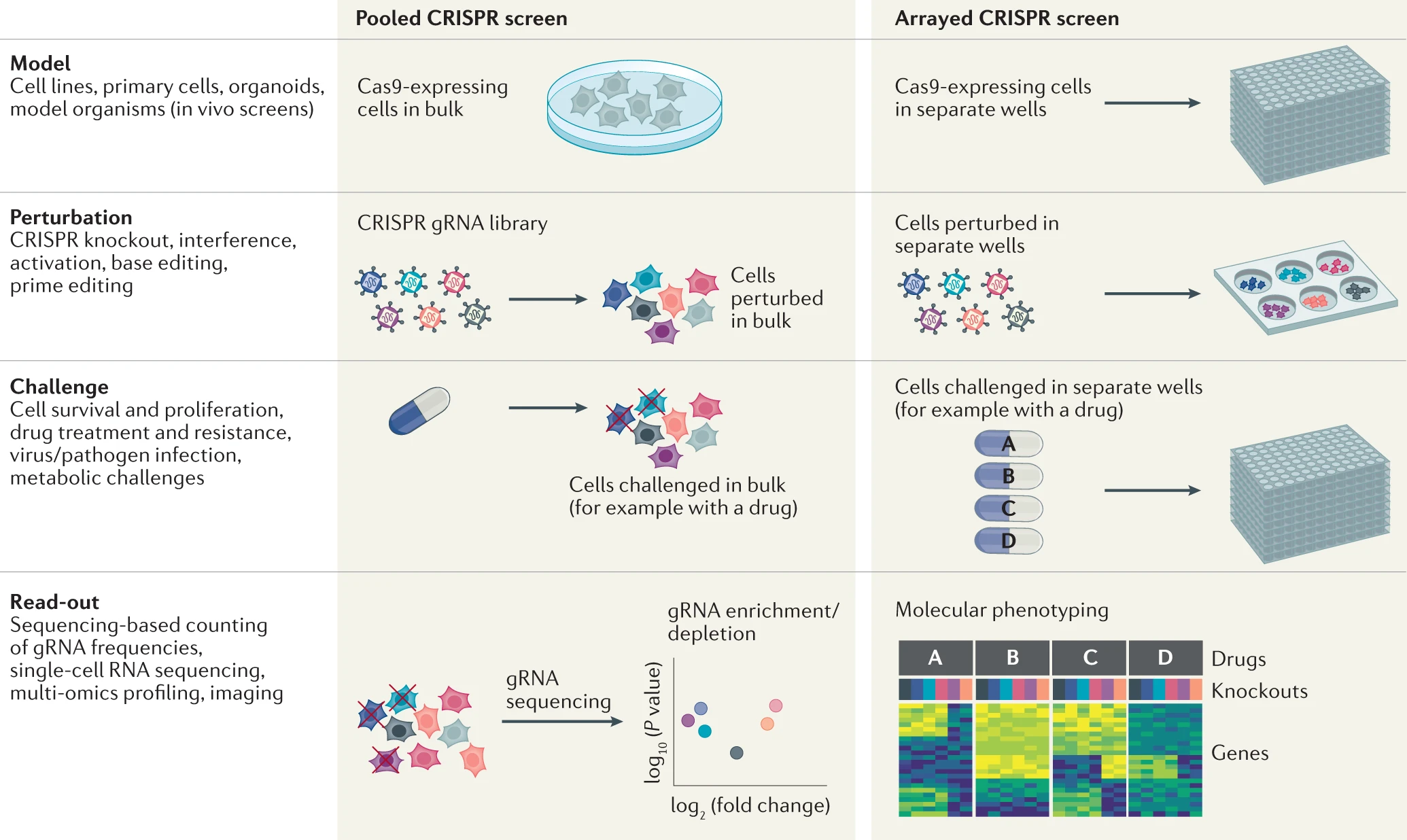
High-content CRISPR screening.
Bock C*, Datlinger P, Chardon F, Coelho MA, Dong MB, Lawson KA, Lu T, Maroc L, Norman TM, Song B, Stanley G, Chen S, Garnett M, Li W, Moffat J, Qi LS, Shapiro RS, Shendure J, Weissman JS, Zhuang X
Nature Reviews Methods Primers 2, 8 (2022). DOI: 10.1038/s43586-021-00093-4
Donwload PDF
Read online (without subscription)
Project website
Press release

Ultra-high-throughput single-cell RNA sequencing and perturbation screening with combinatorial fluidic indexing
Datlinger P, Rendeiro AF#, Boenke T, Senekowitsch M, Krausgruber T, Barreca D, Bock C*
Nature Methods 18, 635-642 (2021). DOI: 10.1038/s41592-021-01153-z
Donwload PDF
Read online (without subscription)

Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden
Peneder P, Stütz AM#, Surdez D, Krumbholz M, Semper S, Chicard M, Sheffield NC, Pierron G, Lapouble E, Totzl M, Erguner B, Barreca D, Rendeiro AF, Agaimy A, Boztug H, Engstler G, Dworzak M, Bernkopf M, Taschner-Mandl S, Ambros IM, Myklebost O, Marec-Bérard P, Burchill SA, Brennan B, Strauss SJ, Whelan J, Schleiermacher G, Schaefer C, Dirksen U, Hutter C, Boye K, Ambros PF, Delattre O, Metzler M, Bock C*, Tomazou EM*
Nature Communications 12, 3230 (2021). DOI: 10.1038/s41467-021-23445-w
Donwload PDF
Read online (without subscription)

The Organoid Cell Atlas
Bock C*, Boutros M, Camp JG, Clarke L, Clevers H, Knoblich JA, Liberali P, Regev A, Rios AC, Stegle O, Stunnenberg HG, Teichmann SA, Treutlein B, Vries RGJ, the Human Cell Atlas ‘Biological Network’ Organoids
Nature Biotechnology 39, 13-17 (2021). DOI: 10.1038/s41587-020-00762-x
Donwload PDF
Read online (without subscription)
Press release

Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2
Popa A, Genger JW#, Nicholson MD, Penz T, Schmid D, Aberle SW, Agerer B, Lercher A, Endler L, Colaco H, Smyth M, Schuster M, Grau ML, Martinez-Jimenez F, Pich O, Borena W, Pawelka E, Keszei Z, Senekowitsch M, Laine J, Aberle JH, Redlberger-Fritz M, Karolyi M, Zoufaly A, Maritschnik S, Borkovec M, Hufnagl P, Nairz M, Weiss G, Wolfinger MT, von Laer D, Superti-Furga G, Lopez-Bigas N, Puchhammer-Stockl E, Aller-berger F, Michor F, Bock C, Bergthaler A*
Science Translational Medicine 12, eabe2555 (2020). DOI: 10.1126/scitranslmed.abe2555
Donwload PDF
Read online (without subscription)

Knowledge-primed neural networks enable biologically interpretable deep learning on single-cell sequencing data
Fortelny N, Bock C*
Genome Biology 21, 190 (2020). DOI: 10.1186/s13059-020-02100-5
Donwload PDF
Read online (without subscription)

Structural cells are key regulators of organ-specific immune response
Krausgruber T, Fortelny N#, Fife-Gernedl V, Senekowitsch M, Schuster LC, Nemc A, Schmidl C, Rendeiro AF, Lercher A, Bergthaler A, Bock C*
Nature 583, 296-302 (2020). DOI: 10.1038/s41586-020-2424-4
Donwload PDF
Read online (without subscription)

Single-cell RNA-seq with spike-in cells enables accurate quantification of cell-specific drug effects in pancreatic islets
Marquina-Sanchez B, Fortelny N#, Farlik M#, Vieira A, Collombat P, Bock C*, Kubicek S*
Genome Biology 21, 106 (2020). DOI: 10.1186/s13059-020-02006-2
Donwload PDF
Read online (without subscription)

Chromatin mapping and single-cell immune profiling defines the temporal dynamics of ibrutinib drug response in chronic lymphocytic leukemia
Rendeiro AF, Krausgruber T#, Fortelny N, Zhao F, Penz T, Farlik M, Schuster LC, Kuchler A, Tasnády S, Réti M, Zoltán M, Alpar D#, Bödör C#, Schmidl C#, Bock C*
Nature Communications 11, 577 (2020). DOI: 10.1038/s41467-019-14081-6
Donwload PDF
Read online (without subscription)

Epigenomics and single-cell sequencing define a developmental hierarchy in Langerhans cell histiocytosis
Halbritter F, Farlik M#, Schwentner R, Jug G, Fortelny N, Schnöller T, Pisa H, Schuster LC, Reinprecht A, Czech T, Gojo J, Holter W, Minkov M, Bauer WM, Simonitsch-Klupp I, Bock C*, Hutter C*
Cancer Discovery 9, 1406-1421 (2019). DOI: 10.1158/2159-8290.CD-19-0138
Donwload PDF
Read online (without subscription)

RnBeads 2.0: comprehensive analysis of DNA methylation data
Müller F, Scherer M#, Assenov Y#, Lutsik P#, Walter J, Lengauer T, Bock C
Genome Biology 20, 55 (2019). DOI: 10.1186/s13059-019-1664-9
Donwload PDF
Read online (without subscription)
Press release

Combined chemosensitivity and chromatin profiling prioritizes drug combinations in CLL
Schmidl C, Vladimer GI#, Rendeiro AF#, Schnabl S#, Krausgruber T, Taubert C, Krall N, Pemovska T, Araghi M, Snijder B, Hubmann R, Ringler A, Runggatscher K, Demirtas D, de la Fuente OL, Hilgarth M, Skrabs C, Porpaczy E, Gruber M, Hoermann G, Kubicek S, Staber PB, Shehata M#, Superti-Furga G#, Jäger U#, Bock C*
Nature Chemical Biology 15, 232-240 (2019). DOI: 10.1038/s41589-018-0205-2
Donwload PDF
Read online (without subscription)

The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space
Klughammer J, Kiesel B#, Roetzer T, Fortelny N, Nemc A, Nenning KH, Furtner J, Sheffield NC, Datlinger P, Peter N, Nowosielski M, Augustin M, Mischkulnig M, Strobel T, Alpar D, Ergüner B, Senekowitsch M, Moser P, Freyschlag CF, Kerschbaumer J, Thomé C, Grams AE, Stockhammer G, Kitzwoegerer M, Oberndorfer S, Marhold F, Weis S, Trenkler J, Buchroithner J, Pichler J, Haybaeck J, Krassnig S, Mahdy Ali K, von Campe G, Payer F, Sherif C, Preiser J, Hauser T, Winkler PA, Kleindienst W, Wurtz F, Brandner-Kokalj T, Stultschnig M, Schweiger S, Dieckmann K, Preusser M, Langs G, Baumann B, Knosp E, Widhalm G, Marosi C, Hainfellner JA, Woehrer A#, Bock C
Nature Medicine 24, 1611-1624 (2018). DOI: 10.1038/s41591-018-0156-x
Donwload PDF
Read online (without subscription)

VCF.Filter: interactive prioritization of disease-linked genetic variants from sequencing data
Müller H*, Jimenez-Heredia R, Krolo A, Hirschmugl T, Dmytrus J, Boztug K, Bock C*
Nucleic Acids Research 45, W567-W572 (2017). DOI: 10.1093/nar/gkx425
Donwload PDF
Read online (without subscription)
Press release

Pooled CRISPR screening with single-cell transcriptome readout
Datlinger P, Schmidl C, Rendeiro A, Krausgruber T, Traxler P, Klughammer J, Schuster L, Kuchler A, Alpar D, Bock C*
Nature Methods 14, 297-301 (2017). DOI: 10.1038/nmeth.4177
Donwload PDF
Read online (without subscription)

DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma
Sheffield NC, Pierron G, Klughammer J, Datlinger P, Schönegger A, Schuster M, Hadler J, Surdez D, Guil-lemot D, Lapouble E, Freneaux P, Champigneulle J, Bouvier R, Walder D, Ambros IM, Hutter C, Sorz E, Am-aral AT, de Alava E, Schallmoser K, Strunk D, Rinner B, Liegl-Atzwanger B, Huppertz B, Leithner A, de Pin-ieux G, Terrier P, Laurence V, Michon J, Ladenstein R, Holter W, Windhager R, Dirksen U, Ambros PF, Delat-tre O, Kovar H, Bock C*, Tomazou EM*
Nature Medicine 23, 386-395 (2017). DOI: 10.1038/nm.4273
Donwload PDF
Read online (without subscription)

DNA methylation dynamics of human hematopoietic stem cell differentiation
Farlik M, Halbritter F#, Müller F#, Choudry FA, Ebert P, Klughammer J, Farrow S, Santoro A, Ciaurro V, Mathur A, Uppal R, Stunnenberg HG, Ouwehand WH, Laurenti E, Lengauer T, Frontini M*, Bock C*
Cell Stem Cell 19, 808-822 (2016). DOI: 10.1016/j.stem.2016.10.019
Donwload PDF
Read online (without subscription)

Preserve personal freedom in networked societies
Bock C
Nature 537, 9 (2016). DOI: 10.1038/537009a
Donwload PDF
Read online (without subscription)
Project website
Press release

Specification of tissue-resident macrophages during organogenesis
Mass E, Ballesteros I#, Farlik M#, Halbritter F#, Günther P, Crozet L, Jacome-Galarza CE, Händler K, Klughammer J, Kobayashi Y, Gomez-Perdiguero E, Schultze JL, Beyer M#, Bock C#, Geissmann F*
Science 353, 6304 (2016). DOI: 10.1126/science.aaf4238
Donwload PDF
Read online (without subscription)

Chromatin accessibility maps of chronic lymphocytic leukaemia identify subtype-specific epigenome signatures and transcription regulatory networks
Rendeiro AF, Schmidl C#, Strefford JC#, Walewska R, Davis Z, Farlik M, Oscier D, Bock C*
Nature Communications 7, 11938 (2016). DOI: 10.1038/ncomms11938
Donwload PDF
Read online (without subscription)

Quantitative comparison of DNA methylation assays for large-scale validation and epigenetic biomarker development
Bock C*, Halbritter F, Carmona FJ, Tierling S, Datlinger P, Assenov Y, Berdasco M, Bergmann AK, Booher K, Busato F, Campan M, Dahl C, Dahmcke CM, Diep D, Fernández AF, Gerhauser C, Haake A, Heilmann K, Hol-comb T, Hussmann D, Ito M, Kreutz M, Kulis M, Lopez V, Nair SS, Paul DS, Plongthongkum N, Qu W, Quei-rós AC, Sauter G, Schlomm T, Stirzaker C, Statham A, Strogantsev R, Urdinguio RG, Walter K, Weichenhan D, Weisenberger DJ, Beck S, Clark SJ, Esteller M, Ferguson-Smith AC, Fraga MF, Guldberg P, Hansen LL, Laird PW, Martin-Subero JI, Nygren AOH, Peist R, Plass C, Shames DS, Siebert R, Sun X, Tost J, Walter J, Zhang K, for the BLUEPRINT consortium
Nature Biotechnology 34, 726-737 (2016). DOI: 10.1038/nbt.3605
Donwload PDF
Read online (without subscription)

Multi-omics of single cells: Strategies and applications
Bock C*, Farlik M, Sheffield NC
Trends in Biotechnology 34, 605-608 (2016). DOI: 10.1016/j.tibtech.2016.04.004
Donwload PDF
Read online (without subscription)
Project website
Press release

Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types
Li J, Klughammer J#, Farlik M#, Penz T#, Spittler A, Barbieux C, Berishvili E, Bock C*, Kubicek S*
EMBO Reports 17, 178-187 (2016). DOI: 10.15252/embr.201540946
Donwload PDF
Read online (without subscription)

LOLA: Enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor
Sheffield NC*, Bock C*
Bioinformatics 32, 587-589 (2016). DOI: 10.1093/bioinformatics/btv612
Donwload PDF
Read online (without subscription)
Press release

Differential DNA methylation analysis without a reference genome
Klughammer J, Datlinger P, Printz D, Sheffield NC, Farlik M, Hadler J, Fritsch G, Bock C*
Cell Reports 13, 2621-2633 (2015). DOI: 10.1016/j.celrep.2015.11.024
Donwload PDF
Read online (without subscription)
Press release

ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors
Schmidl C, Rendeiro AF#, Sheffield NC, Bock C*
Nature Methods 12, 963-965 (2015). DOI: 10.1038/nmeth.3542
Donwload PDF
Read online (without subscription)

Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics
Farlik M, Sheffield NC#, Nuzzo A, Datlinger P, Schönegger A, Klughammer J, Bock C*
Cell Reports 10, 1386-1397 (2015). DOI: 10.1016/j.celrep.2015.02.001
Donwload PDF
Read online (without subscription)

Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1
Tomazou EM, Sheffield NC#, Schmidl C, Schuster M, Schönegger A, Datlinger P, Kubicek S, Bock C*, Kovar H*
Cell Reports 10, 1082-1095 (2015). DOI: 10.1016/j.celrep.2015.01.042
Donwload PDF
Read online (without subscription)

Comprehensive analysis of DNA methylation data with RnBeads
Assenov Y, Müller F#, Lutsik P#, Walter J, Lengauer T, Bock C*
Nature Methods 11, 1138-1140 (2014). DOI: 10.1038/nmeth.3115
Donwload PDF
Read online (without subscription)
Press release

Recommendations for the design and analysis of epigenome-wide association studies
Michels KB*, Binder AM, Dedeurwaerder S, Epstein CB, Greally JM, Gut I, Houseman EA, Izzi B, Kelsey KT, Meissner A, Milosavljevic A, Siegmund KD, Bock C#, Irizarry RA
Nature Methods 10, 949-955 (2013). DOI: 10.1038/nmeth.2632
Read online (without subscription)
Project website
Press release

Analysing and interpreting DNA methylation data
Bock C*
Nature Reviews Genetics 13, 705-719 (2012). DOI: 10.1038/nrg3273
Read online (without subscription)
Project website
Press release

Managing drug resistance in cancer: Lessons from HIV therapy
Bock C*, Lengauer T
Nature Reviews Cancer 12, 494-501 (2012). DOI: 10.1038/nrc3297
Read online (without subscription)
Project website
Press release
Main publications from Christoph Bock’s postdoctoral research (Broad Institute & Harvard University) and PhD studies (Max Planck Institute for Informatics)

EpiExplorer: live exploration and global analysis of large epigenomic datasets
Halachev K*, Bast H, Albrecht F, Lengauer T, Bock C*
Genome Biology 13, R96 (2012). DOI: 10.1186/gb-2012-13-10-r96
Donwload PDF
Read online (without subscription)
Press release

DNA methylation dynamics during in vivo differentiation of blood and skin stem cells
Bock C, Beerman I, Lien WH, Smith Z, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A*
Molecular Cell 47, 633-47 (2012). DOI: 10.1016/j.molcel.2012.06.019
Donwload PDF
Read online (without subscription)
Press release

RRBSMAP: A fast, accurate and user-friendly alignment tool for reduced representation bisulfite sequencing
Xi Y, Bock C#, Müller F, Sun D, Meissner A, Li W*
Bioinformatics 28, 430-432 (2011). DOI: 10.1093/bioinformatics/btr668
Donwload PDF
Read online (without subscription)
Press release

BiQ Analyzer HT: locus-specific analysis of DNA methylation by high-throughput bisulfite sequencing
Lutsik P, Feuerbach L, Arand J, Lengauer T, Walter J*, Bock C*
Nucleic Acids Research 39, W551-556 (2011). DOI: 10.1093/nar/gkr312
Donwload PDF
Read online (without subscription)
Press release

Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines
Bock C, Kiskinis E#, Verstappen G#, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K*, Meissner A*
Cell 144, 439-452 (2011). DOI: 10.1016/j.cell.2010.12.032
Donwload PDF
Read online (without subscription)
Press release

Quantitative comparison of genome-wide DNA methylation mapping technologies
Bock C*, Tomazou EM#, Brinkman AB, Müller F, Simmer F, Gu H, Jäger N, Gnirke A, Stunnenberg HG, Meissner A*
Nature Biotechnology 28, 1106-1114 (2010). DOI: 10.1038/nbt.1681
Donwload PDF
Read online (without subscription)
Press release

Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution
Gu H, Bock C#, Mikkelsen TS, Jäger N, Smith ZD, Tomazou EM, Gnirke A, Lander ES, Meissner A*
Nature Methods 7, 133-136 (2010). DOI: 10.1038/nmeth.1414
Donwload PDF
Read online (without subscription)
Press release

Epigenetic biomarker development
Bock C*
Epigenomics 1, 99-110 (2009). DOI: 10.2217/epi.09.6
Donwload PDF
Read online (without subscription)
Project website
Press release

MethMarker: User-friendly design and optimization of gene-specific DNA methylation assays
Schüffler P, Mikeska T, Waha A, Lengauer T, Bock C*
Genome Biology 10, R105 (2009). DOI: 10.1186/gb-2009-10-10-r105
Donwload PDF
Read online (without subscription)
Press release

EpiGRAPH: User-friendly software for statistical analysis and prediction of epigenomic data
Bock C*, Halachev K, Büch J, Lengauer T
Genome Biology 10, R14 (2009). DOI: 10.1186/gb-2009-10-2-r14
Donwload PDF
Read online (without subscription)
Press release

Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping
Bock C*, Walter J, Paulsen M, Lengauer T
Nucleic Acids Research 36, e55 (2008). DOI: 10.1093/nar/gkn122
Donwload PDF
Read online (without subscription)
Project website
Press release

Computational epigenetics
Bock C*, Lengauer T
Bioinformatics 24, 01.Oct (2008). DOI: 10.1093/bioinformatics/btm546
Donwload PDF
Read online (without subscription)
Project website
Press release

CpG island mapping by epigenome prediction
Bock C*, Walter J, Paulsen M, Lengauer T
PLOS Computational Biology 3, e110 (2007). DOI: 10.1371/journal.pcbi.0030110
Donwload PDF
Read online (without subscription)
Project website
Press release

CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure
Bock C*, Paulsen M, Tierling S, Mikeska T, Lengauer T, Walter J
PLOS Genetics 2, e26 (2006). DOI: 10.1371/journal.pgen.0020026
Donwload PDF
Read online (without subscription)
Project website
Press release

BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing
Bock C*, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T
Bioinformatics 21, 4067-4068 (2005). DOI: 10.1093/bioinformatics/bti652
Donwload PDF
Read online (without subscription)
Press release