High-throughput biology of cancer and immunity

We use multi-omics profiling and CRISPR technology to discover exciting new biology in cancer and immunity, and we take the most interesting results forward, together with our clinical partners to make them useful for precision medicine.
The biological concept of cellular identity connects a cell’s past with its future potential. Our research is based on the hypothesis that epigenetic cell states and the “epigenetic landscape” constitute a highly informative intermediate layer of information processing – one that captures a cell’s regulatory identity and retains the flexibility to respond swiftly to a broad range of perturbations. This definition of the “epigenetic landscape” is not restricted to specific epigenetic marks such as DNA methylation and histone modifications. Rather, it reflects the full range of transcription regulation by which cells translate various inputs into sustainable changes in their cell state. In particular, the epigenetic landscape not only defines a cell’s current state, but also details its developmental past (e.g., cell-of-origin in cancer) and future trajectories (e.g., plasticity in response to drug treatment).
To study the fundamental role of epigenetic and transcription-regulatory programs in cancer and immunity, we have established high-throughput pipelines to profile gene expression (i.e., bulk and single-cell RNA-seq) and transcriptional regulation (e.g., ATAC-seq, RRBS, ChIPmentation) to dissect the epigenetic landscape in patient cohorts and in disease-relevant mouse models. For example, we have established genome-scale maps of tumor heterogeneity in cancers of children (Ewing sarcoma, Langerhans histiocytosis) and elderly patients (glioblastoma, chronic lymphocytic leukemia). Our work also focuses on immune gene regulation – the first single-cell DNA methylation maps of hematopoietic stem cell differentiation and the first systematic map of immune regulation in non-immune cells (“structural immunity”).
Publications

Integrated time-series analysis and high-content CRISPR screening delineate the dynamics of macrophage immune regulation
Traxler P, Reichl S#, Folkman L, Shaw L, Fife V, Nemc A, Pasajlic DJ, Kusienicka A, Barreca D, Fortelny N, Rendeiro AF, Halbritter F, Weninger W, Decker T, Farlik M#, Bock C*
Cell Systems (2025). DOI: 10.1016/j.cels.2025.101346
Donwload PDF
Read online (without subscription)

GPT-4 as a biomedical simulator
Schaefer M, Reichl S#, ter Horst R#, Nicolas AM, Krausgruber T, Piras F, Stepper P, Bock C*, Samwald M*
Computers in Biology and Medicine 178, 108796 (2024). DOI: 10.1016/j.compbiomed.2024.108796
Donwload PDF
Read online (without subscription)
Press release
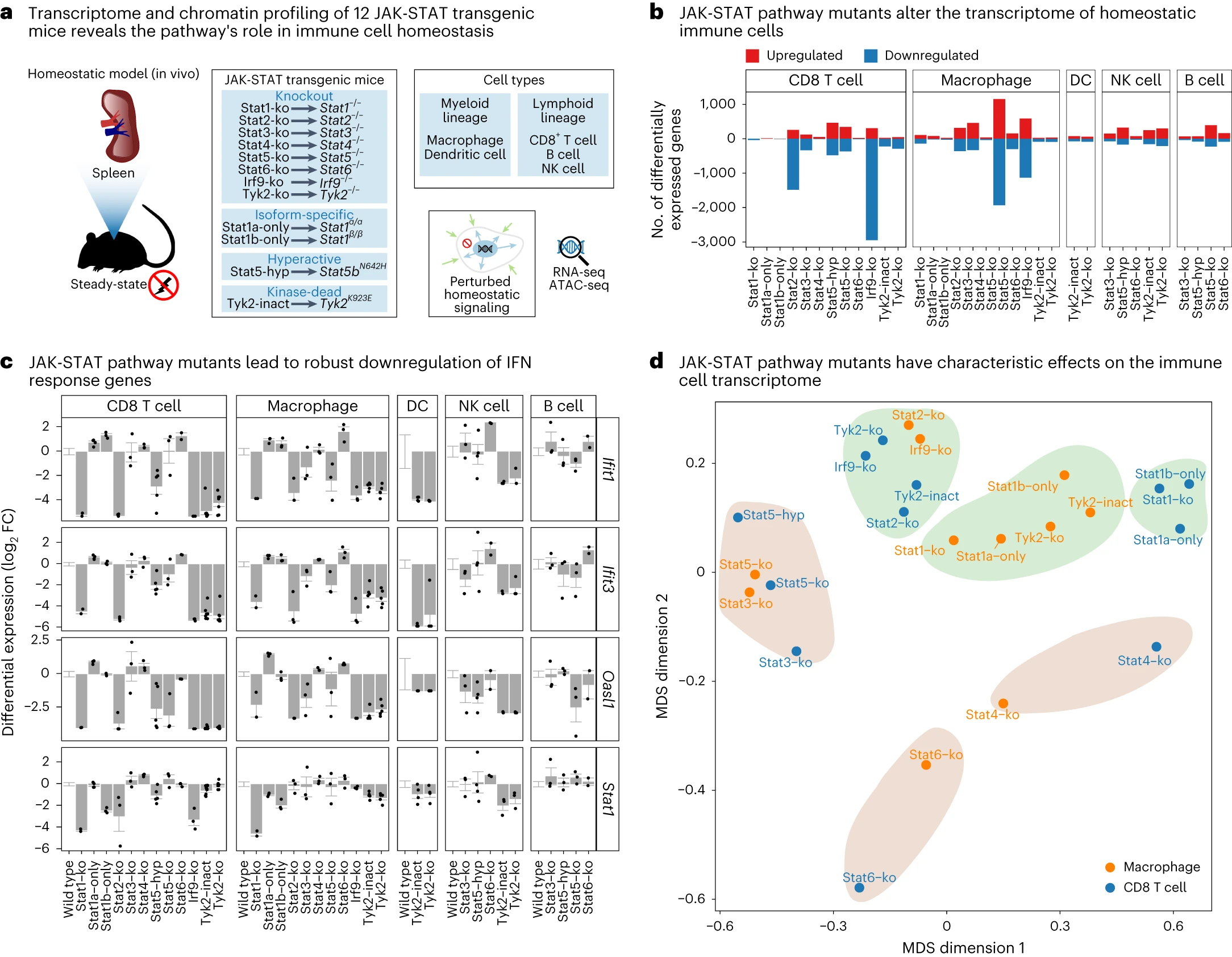
JAK-STAT signaling maintains homeostasis in T cells and macrophages
Fortelny N, Farlik M#*, Fife V, Gorki AD, Lassnig C, Maurer B, Meissl K, Dolezal M, Boccuni L, Ravi Sundar Jose Geetha A, Akagha MJ, Karjalainen A, Shoebridge S, Farhat A, Mann U, Jain R, Tikoo S, Zila N, Esser-Skala W, Krausgruber T, Sitnik K, Penz T, Hladik A, Suske T, Zahalka S, Senekowitsch M, Barreca D, Halbritter F, Macho-Maschler S, Weninger W, Neubauer HA, Moriggl R, Knapp S, Sexl V, Strobl B, Decker T, Müller M, Bock C*
Nature Immunology 25 (2024). DOI: 10.1038/s41590-024-01804-1
Donwload PDF
Read online (without subscription)

Multi-omics analysis of innate and adaptive responses to BCG vaccination reveals epigenetic cell states that predict trained immunity
Moorlag SJCFM, Folkman L#, ter Horst R#, Krausgruber T#, Barreca D, Schuster LC, Fife V, Matzaraki V, Li W, Reichl S, Mourits VP, Koeken VACM, de Bree CJ, Dijkstra H, Lemmers H, van Cranenbroek B, van Rijssen E, Koenen HJP, Jossten I, Xu CJ, Li Y, Joosten LAB, van Crevel R, Netea M*, Bock C*
Immunity 57/1, 171-187 (2024). DOI: 10.1016/j.immuni.2023.12.005
Donwload PDF
Read online (without subscription)
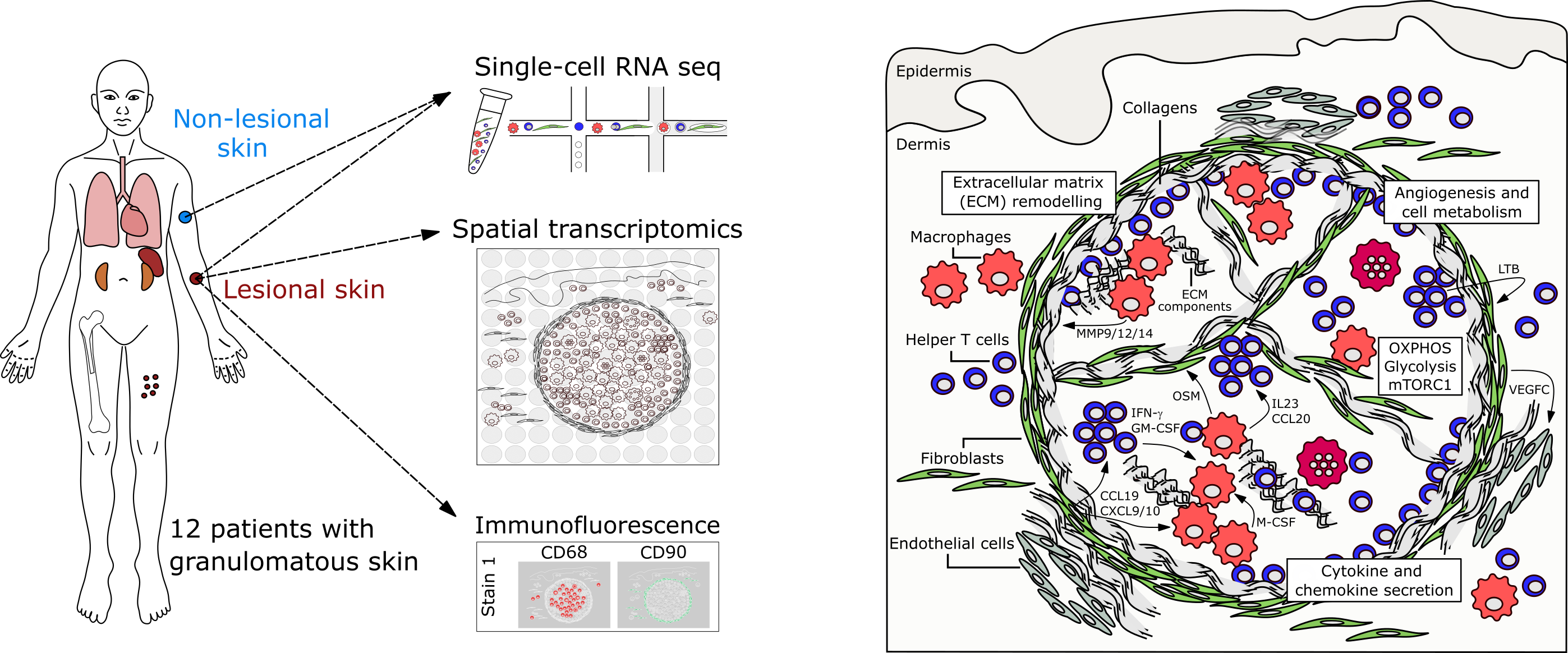
Single-cell and spatial transcriptomics reveal aberrant lymphoid developmental programs driving granuloma formation
Krausgruber T, Redl A#, Barreca D#, Doberer K, Romanovskaia D, Dobnikar L, Guarini M, Unterluggauer L, Kleissl L, Atzmüller D, Mayerhofer C, Kopf A, Saluzzo S, Lim CX, Rexie P, Weichhart T, Bock C*, Stary G*
Immunity 56, 289-306 (2023). DOI: 10.1016/j.immuni.2023.01.014
Donwload PDF
Read online (without subscription)

Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden
Peneder P, Stutz AM, Surdez D, Krumbholz M, Semper S, Chicard M, Sheffield NC, Pierron G, Lapouble E, Totzl M, Erguner B, Barreca D, Rendeiro AF, Agaimy A, Boztug H, Engstler G, Dworzak M, Bernkopf M, Taschner-Mandl S, Ambros IM, Myklebost O, Marec-Berard P, Burchill SA, Brennan B, Strauss SJ, Whelan J, Schleiermacher G, Schaefer C, Dirksen U, Hutter C, Boye K, Ambros PF, Delattre O, Metzler M, Bock C, Tomazou EM
Nature Communications 12, 3230 (2021). DOI: 10.1038/s41467-021-23445-w
Donwload PDF
Read online (without subscription)

Structural cells are key regulators of organ-specific immune response
Krausgruber T*, Fortelny N*, Fife-Gernedl V, Senekowitsch M, Schuster LC, Nemc A, Schmidl C, Rendeiro AF, Lercher A, Bergthaler A, Bock C
Nature 583, 296-302 (2020). DOI: 10.1038/s41586-020-2424-4
Read online (without subscription)

Chromatin mapping and single-cell immune profiling defines the temporal dynamics of ibrutinib drug response in chronic lymphocytic leukemia
Rendeiro AF*, Krausgruber T*, Fortelny N, Zhao F, Penz T, Farlik M, Schuster LC, Kuchler A, Tasnády S, Réti M, Zoltán M, Alpar D*, Bödör C*, Schmidl C*, Bock C*
Nature Communications 11, 577 (2020). DOI: 10.1038/s41467-019-14081-6
Donwload PDF
Read online (without subscription)

Epigenomics and single-cell sequencing define a developmental hierarchy in Langerhans cell histiocytosis
Halbritter F*, Farlik M*, Schwentner R, Jug G, Fortelny N, Schnöller T, Pisa H, Schuster LC, Reinprecht A, Czech T, Gojo J, Holter W, Minkov M, Bauer WM, Simonitsch-Klupp I, Bock C*, Hutter C*
Cancer Discovery 9, 1406-1421 (2019). DOI: 10.1158/2159-8290.CD-19-0138
Donwload PDF
Read online (without subscription)

Combined chemosensitivity and chromatin profiling prioritizes drug combinations in CLL
Schmidl C*, Vladimer GI*, Rendeiro AF*, Schnabl S*, Krausgruber T, Taubert C, Krall N, Pemovska T, Araghi M, Snijder B, Hubmann R, Ringler A, Runggatscher K, Demirtas D, de la Fuente OL, Hilgarth M, Skrabs C, Porpaczy E, Gruber M, Hoermann G, Kubicek S, Staber PB, Shehata M*, Superti-Furga G*, Jäger U*, Bock C*
Nature Chemical Biology 15, 232-240 (2019). DOI: 10.1038/s41589-018-0205-2
Donwload PDF
Read online (without subscription)

The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space
Klughammer J*, Kiesel B*, Roetzer T, Fortelny N, Nemc A, Nenning KH, Furtner J, Sheffield NC, Datlinger P, Peter N, Nowosielski M, Augustin M, Mischkulnig M, Strobel T, Alpar D, Ergüner B, Senekowitsch M, Moser P, Freyschlag CF, Kerschbaumer J, Thomé C, Grams AE, Stockhammer G, Kitzwoegerer M, Oberndorfer S, Marhold F, Weis S, Trenkler J, Buchroithner J, Pichler J, Haybaeck J, Krassnig S, Mahdy Ali K, von Campe G, Payer F, Sherif C, Preiser J, Hauser T, Winkler PA, Kleindienst W, Wurtz F, Brandner-Kokalj T, Stultschnig M, Schweiger S, Dieckmann K, Preusser M, Langs G, Baumann B, Knosp E, Widhalm G, Marosi C, Hainfellner JA, Woehrer A*, Bock C*
Nature Medicine 24, 1611-1624 (2018). DOI: 10.1038/s41591-018-0156-x
Donwload PDF
Read online (without subscription)

DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma
Sheffield NC, Pierron G, Klughammer J, Datlinger P, Schönegger A, Schuster M, Hadler J, Surdez D, Guillemot D, Lapouble E, Freneaux P, Champigneulle J, Bouvier R, Walder D, Ambros IM, Hutter C, Sorz E, Amaral AT, de Alava E, Schallmoser K, Strunk D, Rinner B, Liegl-Atzwanger B, Huppertz B, Leithner A, de Pinieux G, Terrier P, Laurence V, Michon J, Ladenstein R, Holter W, Windhager R, Dirksen U, Ambros PF, Delattre O, Kovar H, Bock C*, Tomazou EM*
Nature Medicine 23, 386-395 (2017). DOI: 10.1038/nm.4273
Donwload PDF
Read online (without subscription)

Specification of tissue-resident macrophages during organogenesis
Mass E*, Ballesteros I*, Farlik M*, Halbritter F*, Günther P, Crozet L, Jacome-Galarza CE, Händler K, Klughammer J, Kobayashi Y, Gomez-Perdiguero E, Schultze JL, Beyer M*, Bock C*, Geissmann F*
Science 353, 6304 (2016). DOI: 10.1126/science.aaf4238
Donwload PDF
Read online (without subscription)

Chromatin accessibility maps of chronic lymphocytic leukaemia identify subtype-specific epigenome signatures and transcription regulatory networks
Rendeiro AF*, Schmidl C*, Strefford JC*, Walewska R, Davis Z, Farlik M, Oscier D, Bock C*
Nature Communications 7, 11938 (2016). DOI: 10.1038/ncomms11938
Donwload PDF
Read online (without subscription)
* shared first or shared senior authorship


