New technologies for high-throughput biology

Important biological discoveries are often enabled by groundbreaking new technologies. We are strongly committed to technology development, especially in the areas of epigenome profiling, single-cell sequencing, genome/epigenome editing, and synthetic biology.
Epigenome profiling. We have developed a series of methods for low-input and single-cell epigenome profiling, which includes ChIPmentation for histone marks and single-cell whole genome bisulfite sequencing (scWGBS) for DNA methylation. Moreover, we have established robust end-to-end pipelines for DNA methylation profiling (using an optimized RRBS protocol) and for chromatin accessibility profiling (using an optimized ATAC-seq protocol), with which we have processed several thousand patient samples each, in numerous projects and collaborations.
Single-cell sequencing. Our lab has been an early adopter of various single-cell sequencing technologies including Smart-seq, Drop-seq, and 10x Genomics profiling. Moreover, we have recently developed the scifi-RNA-seq method, which combines combinatorial indexing with droplet microfluidics for single-cell RNA-seq in millions of cells and thousands of samples.
CRISPR screening. We have developed the CROP-seq technology for CRISPR screening with single-cell transcriptome readout. This method enables functional biology at scale, with the potential of accelerating biological discovery and clinical translation. CROP-seq combines the scalability of pooled CRISPR screening with single-cell transcriptome sequencing as its readout, making it possible to screen for complex regulatory phenotypes.
Collectively, these methods – and additional new methods that are currently under development in our lab – enable us to study biological processes in high resolution and high throughput, in order to home in on disease-relevant mechanisms.
Publications
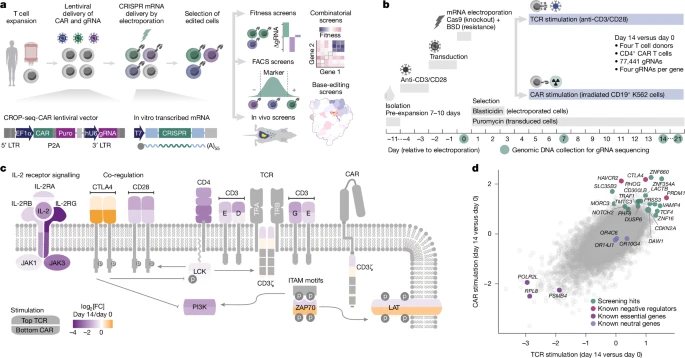
Systematic discovery of CRISPR-boosted CAR T cell immunotherapies
Datlinger P#*, Pankevich EV#, Arnold CD#, Pranckevicius N, Lin J, Romanovskaia D, Schaefer M, Piras F, Orts AC, Nemc A, Biesaga PN, Chan M, Neuwirth T, Artemov AV, Wentao Li, Ladstätter S, Krausgruber T. & Bock C*
Nature (2025). DOI: 10.1038/s41586-025-09507-9
Donwload PDF
Read online (without subscription)
Project website

Integrated time-series analysis and high-content CRISPR screening delineate the dynamics of macrophage immune regulation
Traxler P, Reichl S#, Folkman L, Shaw L, Fife V, Nemc A, Pasajlic DJ, Kusienicka A, Barreca D, Fortelny N, Rendeiro AF, Halbritter F, Weninger W, Decker T, Farlik M#, Bock C*
Cell Systems (2025). DOI: 10.1016/j.cels.2025.101346
Donwload PDF
Read online (without subscription)

Ultra-high-throughput single-cell RNA sequencing and perturbation screening with combinatorial fluidic indexing
Datlinger P, Rendeiro AF, Boenke T, Senekowitsch M, Krausgruber T, Barreca D, Bock C
Nature Methods 18, 635-642 (2021). DOI: 10.1038/s41592-021-01153-z
Donwload PDF
Read online (without subscription)

Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden
Peneder P, Stutz AM, Surdez D, Krumbholz M, Semper S, Chicard M, Sheffield NC, Pierron G, Lapouble E, Totzl M, Erguner B, Barreca D, Rendeiro AF, Agaimy A, Boztug H, Engstler G, Dworzak M, Bernkopf M, Taschner-Mandl S, Ambros IM, Myklebost O, Marec-Berard P, Burchill SA, Brennan B, Strauss SJ, Whelan J, Schleiermacher G, Schaefer C, Dirksen U, Hutter C, Boye K, Ambros PF, Delattre O, Metzler M, Bock C, Tomazou EM
Nature Communications 12, 3230 (2021). DOI: 10.1038/s41467-021-23445-w
Donwload PDF
Read online (without subscription)

Single-cell RNA-seq with spike-in cells enables accurate quantification of cell-specific drug effects in pancreatic islets
Marquina-Sanchez B*, Fortelny N*, Farlik M*, Vieira A, Collombat P, Bock C*, Kubicek S*
Genome Biology 21, 106 (2020). DOI: 10.1186/s13059-020-02006-2
Donwload PDF
Read online (without subscription)

Pooled CRISPR screening with single-cell transcriptome readout
Datlinger P, Schmidl C, Rendeiro A, Krausgruber T, Traxler P, Klughammer J, Schuster L, Kuchler A, Alpar D, Bock C
Nature Methods 14, 297-301 (2017). DOI: 10.1038/nmeth.4177
Donwload PDF
Read online (without subscription)

Quantitative comparison of DNA methylation assays for large-scale validation and epigenetic biomarker development
Bock C, Halbritter F, Carmona FJ, Tierling S, Datlinger P, Assenov Y, Berdasco M, Bergmann AK, Booher K, Busato F, Campan M, Dahl C, Dahmcke CM, Diep D, Fernández AF, Gerhauser C, Haake A, Heilmann K, Holcomb T, Hussmann D, Ito M, Kreutz M, Kulis M, Lopez V, Nair SS, Paul DS, Plongthongkum N, Qu W, Queirós AC, Sauter G, Schlomm T, Stirzaker C, Statham A, Strogantsev R, Urdinguio RG, Walter K, Weichenhan D, Weisenberger DJ, Beck S, Clark SJ, Esteller M, Ferguson-Smith AC, Fraga MF, Guldberg P, Hansen LL, Laird PW, Martin-Subero JI, Nygren AOH, Peist R, Plass C, Shames DS, Siebert R, Sun X, Tost J, Walter J, Zhang K, for the BLUEPRINT consortium
Nature Biotechnology 34, 726-737 (2016). DOI: 10.1038/nbt.3605
Donwload PDF
Read online (without subscription)

Multi-omics of single cells: Strategies and applications
Bock C, Farlik M, Sheffield NC
Trends in Biotechnology 34, 605-608 (2016). DOI: 10.1016/j.tibtech.2016.04.004
Donwload PDF
Read online (without subscription)
Project website
Press release

Differential DNA methylation analysis without a reference genome
Klughammer J, Datlinger P, Printz D, Sheffield NC, Farlik M, Hadler J, Fritsch G, Bock C
Cell Reports 13, 2621-2633 (2015). DOI: 10.1016/j.celrep.2015.11.024
Donwload PDF
Read online (without subscription)
Press release

ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors
Schmidl C*, Rendeiro AF*, Sheffield NC, Bock C
Nature Methods 12, 963-965 (2015). DOI: 10.1038/nmeth.3542
Donwload PDF
Read online (without subscription)

Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics
Farlik M*, Sheffield NC*, Nuzzo A, Datlinger P, Schönegger A, Klughammer J, Bock C
Cell Reports 10, 1386-1397 (2015). DOI: 10.1016/j.celrep.2015.02.001
Donwload PDF
Read online (without subscription)

Analysing and interpreting DNA methylation data
Bock C
Nature Reviews Genetics 13, 705-719 (2012). DOI: 10.1038/nrg3273
Read online (without subscription)
Project website
Press release
* shared first or shared senior authorship